宜宾网站建设哪家好软件开发合同范本大全
腾讯云轻量应用服务器性能评测,轻量服务器CPU主频、处理器型号、公网带宽、月流量、Ping值测速、磁盘IO读写及使用限制,轻量应用服务器CPU内存性能和标准型云服务器CVM处于同一水准,所以大家不要担心轻量应用服务器的性能,腾讯云百科分享腾讯云轻量应用服务器性能测评:
目录
腾讯云轻量应用服务器性能评测
1、轻量CPU处理器型号主频
2、CPU内存计算性能
3、轻量服务器Ping值测速
4、轻量月流量
5、公网带宽
6、轻量服务器系统盘存储
7、轻量应用服务器限制
腾讯云轻量应用服务器性能评测
轻量应用服务器是腾讯云推出的一款开箱即用的轻量级的云服务器,轻量服务器CPU内存带宽配置更高,价格却又很便宜,很多同学认为是不是轻量应用服务器性能不行呀,轻量服务器和云服务器有什么区别?本文云服务器吧主要分享腾讯云轻量应用服务器性能评测,也可以参考官方页面:https://curl.qcloud.com/8Eps6xac
1、轻量CPU处理器型号主频
轻量应用服务器CPU主频是多少?轻量处理器型号是哪款?云服务器吧在腾讯云官网找到了答案,轻量应用服务器并指定CPU型号,用户在创建轻量服务器时,腾讯云系统自动分配的处理器,CPU一般采用较新代次的CPU型号。如下图:

轻量服务器不支持指定底层物理服务器的CPU型号
创建轻量应用服务器时不支持指定底层物理服务器的CPU型号,腾讯云将随机分配满足套餐规格的物理CPU型号,通常优先选择较新代次的CPU型号。相对于CVM云服务器轻量服务器更适合轻量级的应用,轻量服务适合中小企或个人开发者用于搭建We网站应用、小程序、APP以及开发测试环境等,云服务器CVM适用于高并发网站、复杂分布式集群应用、大数据分析等架构复杂的使用场景。关于轻量和CVM区别可以参考:腾讯云轻量服务器和CVM云服务器区别全方位对比(2023更新)
云服务器吧的腾讯云账号下有多台轻量应用服务器,目前采用的CPU有2.5GHz的Intel(R) Xeon(R) Gold 6133 CPU,睿频 3.0GHz,也有2.4GHz主频Intel(R) Xeon(R) CPU E5-26xx v4处理器,如下图:

轻量服务器CPU处理器型号主频
2、CPU内存计算性能
关于轻量应用服务器CPU内存的计算性能,云服务器吧去翻阅官方文档,轻量服务器与同规格的 标准型云服务器CVM相比,轻量应用服务器的CPU、内存性能与其处于同一水准。所以大家不用担心,轻量服务器的计算性能。
3、轻量服务器Ping值测速
腾讯云轻量应用服务器分为中国大陆地域,如北京、上海及广州等地域,在腾讯云多线BGP的加持下,各个节点速度大家完全不用担心。所以云服务器吧认为中国大陆地域的轻量服务器没必要担心,都挺快的,云服务器吧来测试下中国香港地域的轻量应用服务器,很多同学单位香港轻量服务器国内访问会不会绕路,云服务器吧测试了一下,国内电信移动联通三网直连,网络延迟可以参考下表:
| 线路 | 最快节点 | 最慢节点 | 平均响应 |
|---|---|---|---|
| 电信 | 9ms | 100ms | 40.2ms |
| 多线 | 11ms | 215ms | 69.3ms |
| 联通 | 20ms | 114ms | 66.8ms |
| 移动 | 32ms | 53ms | 40ms |
| 海外 | 2ms | 230ms | 50.7ms |
4、轻量月流量
腾讯云轻量应用服务器限制月流量的,轻量服务器是按套餐售卖的,每台轻量服务器都有流量包限制,每个月流量包内产生的流量是免费的,超出月流量包的部分需要另外支付流量费,轻量服务器地域不同流量价格不同,如下表:
| 地域 | 价格(元/GB) |
|---|---|
| 中国内地、新加坡、东京、法兰克福、首尔 | 0.8 |
| 中国香港 | 1.0 |
| 孟买 | 0.58 |
| 硅谷 | 0.5 |
需要注意的是,腾讯云只统计轻量服务器公网出方向的流量,只有公网出方向产生的流量是计费的,公网入方向和私网产生的流量都是免费的,如下图:

腾讯云轻量服务器流量收费和免费说明
更多关于流量计费,请参考:腾讯云轻量应用服务器月流量用完怎么办?收费吗?
5、公网带宽
轻量服务器套餐公网带宽很高,例如2核2G3M、2核2G4M、2核4G5M、4核8G12M、8核16G18M、16核32G28M,而云服务器CVM一般标配1M公网带宽,看起来轻量服务器公网带宽更高,但是实际可能是峰值带宽,并不是永远能够达到固定带宽值,而云服务器CVM的公网带宽是独享的,但是CVM带宽也要更贵一些。

腾讯云轻量服务器公网带宽
综上,轻量应用服务器的公网带宽可能不如云服务器CVM,但是总体性价比方面,轻量应用服务器还是要更胜一筹。
6、轻量服务器系统盘存储
腾讯云轻量应用服务器系统盘全系标配SSD盘,磁盘IO读写如下图所示:
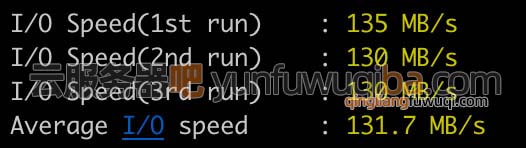
轻量服务器磁盘IO读写
7、轻量应用服务器限制
轻量应用服务器相对于云服务器CVM是有一些限制的,购买前云服务器吧建议大家了解一下:
- 1、轻量应用服务器实例创建完成后,不支持更换内网IP地址,之前公网IP地址也不支持更换,现在可以更换公网IP地址了;
- 2、轻量应用服务器CPU内存带宽系统盘是一个整体套餐,只能整体升级,并且不支持降级套餐;
- 3、不支持用户自定义配置私有网络 VPC,网络由系统自动创建并分配;
- 4、每个账号在单地域下可创建的数据盘限制20块,每台轻量应用服务器实例可同时挂载的数据盘5块;
- 5、每台轻量应用服务器可创建的防火墙规则数量限制为100个;
- 6、每个账号在单地域可创建的SSH密钥对数量限制10个;
- 7、每个地域的自定义镜像配额20个;可领券:https://curl.qcloud.com/HmjGZiLu 服务器专享满减优惠券;
- 8、每个地域内的免费快照总数量上限为已创建实例数(不包含待回收实例及使用存储型套餐的实例)乘以2,且最多不超过10个,使用存储型套餐的实例,不支持创建快照;
- 9、每台轻量应用服务器实例备案的网站不超过5个,每台轻量应用服务器可生成2个备案授权码(仅支持企业账号)。
以上是腾讯云百科分享的腾讯云轻量应用服务器性能测评,包括CPU型号、CPU主频、CPU内存计算性能、公网带宽、月流量包、系统盘存储、网络测速及限制说明,转载请注明云服务器吧。
原文:https://www.bidianbao.com/2656.html
