企业管理网站模板官方网站建设 找磐石网络一流
文章目录
- Nginx产生的原因
- Nginx简介
- Nginx的作用
- 反向代理
- 负载均衡策略
- 动静分离
- Nginx的
- Windows下的安装
- Linux下的安装
- Nginx常用命令
- 负载均衡功能演示
Nginx产生的原因
背景
一个公司的项目刚刚上线的时候,并发量小,用户使用的少,所以在低并发的情况下,一个jar包启动应用就够了,然后内部tomcat返回内容给用户

慢慢的,使用平台的用户越来越多了,并发量慢慢增大了,这时候一台服务器满足不了我们的需求了
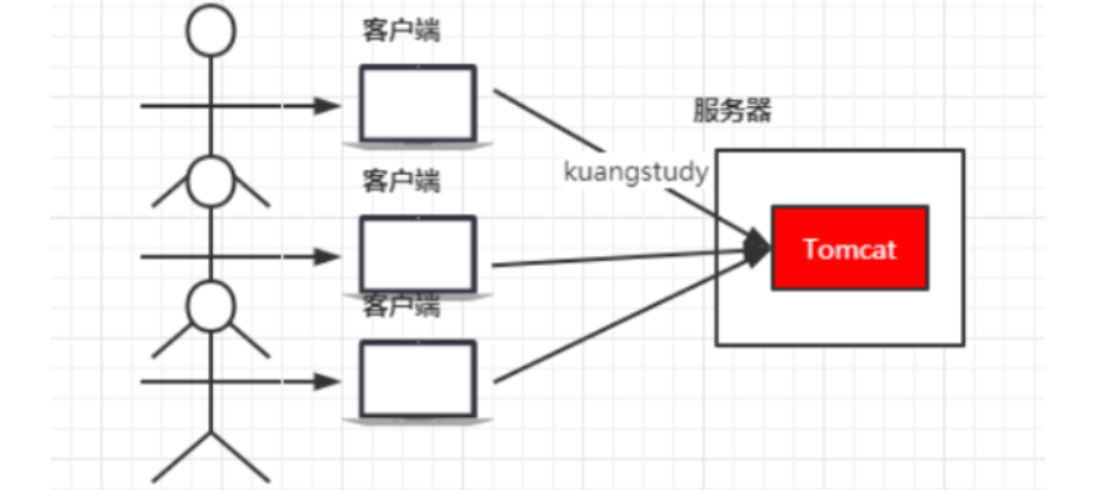
于是我们横向扩展,又增加了服务器。这个时候几个项目启动在不同的服务器上,用户要访问,就需要增加一个代理服务器了,通过代理服务器来帮我们转发和处理请求(由该代理服务器作负载均衡&&反向代理)
- 希望这个代理服务器可以帮助我们接收用户的请求,然后将用户的请求按照规则帮我们转发到不同的服务器节点之上
- 这个过程用户是无感知的,用户并不知道是哪个服务器返回的结果,我们还希望他可以按照服务器的性能提供不同的权重选择,保证最佳体验,基于上述的需求,所以使用了Nginx。

Nginx简介
- Nginx (engine x) 是一个高性能的HTTP和反向代理web服务器,同时也提供了IMAP/POP3/SMTP服务
- 其特点是占有内存少,并发能力强,事实上nginx的并发能力在同类型的网页服务器中表现较好
- Nginx 是一个安装非常的简单、配置文件非常简洁(还能够支持perl语法=>可以理解为正则表达式 )、Bug非常少的服务,Nginx 启动容易,并且几乎可以做到7*24小时不间断运行,即使运行数个月也不需要重新启动,还能够不间断服务的情况下进行软件版本的升级
- Nginx底层是C语言实现的,官方数据测试表明能够支持高达 5w个并发连接数的响应
Nginx的作用
反向代理
- 作为Http代理=>反向代理:作为web服务器最常用的功能之一
关于正向代理和反向代理
正向代理:用于代理客户端的请求(帮客户端请求外部资源),服务器只知道有客户端的请求到来,但是并不知道是哪个客户端请求的 (例如:VPN)
正向代理的工作方式
1.客户端配置网络设置,将请求发送给正向代理服务器
2.正向代理服务器接收到请求后,根据客户端请求的目标地址,代表客户端向目标服务器发送请求
3.目标服务器处理请求并发送响应
4.正向代理服务器接收到响应后,将其返回给客户端

正向代理的主要功能包括:
1.访问控制:正向代理可以根据特定的策略和规则,对客户端的请求进行访问控制,例如限制特定网站的访问或实施身份验证
2.隐私保护:正向代理可以隐藏客户端的真实身份和位置,提供一定的匿名性
3.缓存:正向代理可以缓存常用的内容,以减少网络流量和提高响应速度
4.加速访问:正向代理服务器可以通过压缩、缓存和优化网络连接等方式,加速客户端与目标服务器之间的通信
反向代理 :扮演服务器的角色,接收来自客户端的请求,并将其转发到后端服务器,对于用户是无感知的,客户发送请求,但是不知道哪个服务端收到了请求,客户端只和反向代理进行通信
- 比如,百度的服务器是在不同的服务器上,但是我们只需要搜索baidu.com就行,该请求可能会被反向代理路由到离你物理位置最近的服务器,或者根据网络条件和负载均衡算法进行分配。这样做可以减少响应时间并提高搜索的效率
反向代理的工作方式
1.客户端向反向代理发送请求
2.反向代理接收到请求后,根据预先设置的规则,将请求转发到一个或多个后端服务器
3.后端服务器处理请求并发送响应
4.反向代理接收到响应后,将其返回给客户端
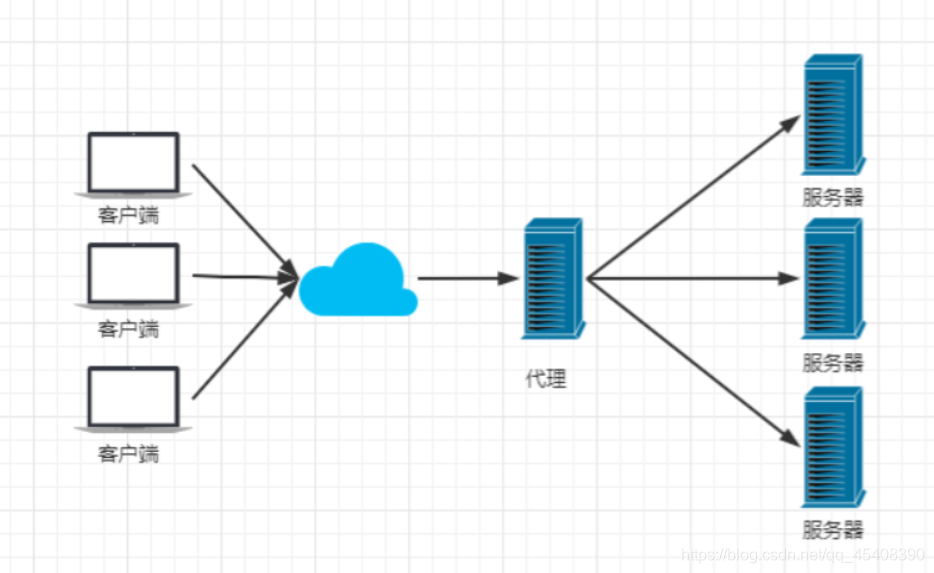
反向代理的主要功能包括
1.负载均衡:反向代理可以将请求分发到多个后端服务器,以实现负载均衡,提高系统的性能和可扩展性。
2.缓存:反向代理可以缓存静态内容,减轻后端服务器的负载,提高响应速度
3.安全性:反向代理可以作为防火墙,保护后端服务器免受恶意请求和攻击
4.SSL 加密:反向代理可以终止 SSL 连接,解密传入的请求,并将其转发给后端服务器,提供安全的通信
负载均衡策略
Nginx提供的负载均衡策略有2种:内置策略和扩展策略
-
内置策略:
轮询,加权轮询,Ip hash- 轮询(Round Robin):默认的负载均衡策略,按照顺序将请求依次分发给后端服务器‘
- IP 哈希(IP Hash):根据客户端的 IP 地址将请求分发给后端服务器,确保同一客户端的请求始终发送到同一台服务器,以保持会话的一致性
- 最少连接(Least Connections):将请求发送给当前连接数最少的后端服务器,以实现负载均衡
- 加权轮询(Weighted Round Robin):根据后端服务器的权重设置,按比例分配请求
- 加权最少连接(Weighted Least Connections):根据后端服务器的权重和当前连接数,动态地分配请求
-
扩展策略:天马行空,只有你想不到的没有他做不到的,可以在
nginx的配置文件当中进行配置
方式1:普通轮询
每个服务器之间轮询处理请求,每台服务器处理请求的数量和概率都是相同的

方式2:加权轮询:因为可能不同服务器性能不一样,能接收的请求数量不同
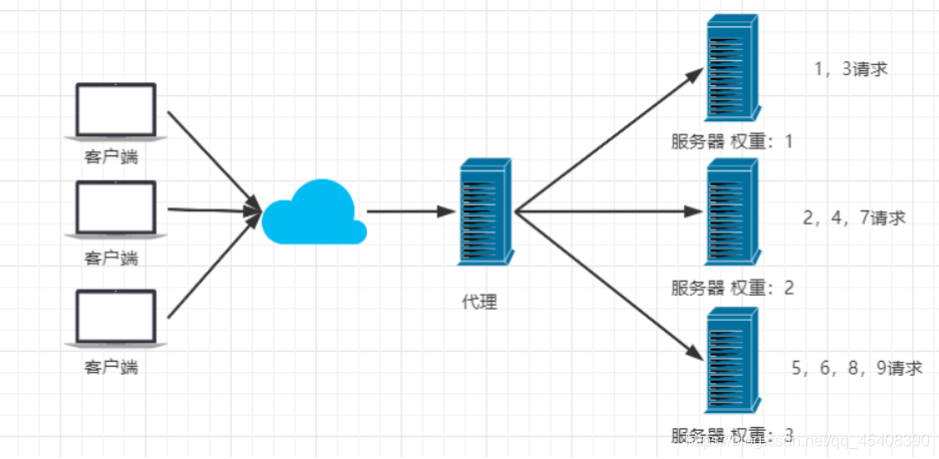
方式3:IP Hash
对客户端请求的ip进行hash操作,然后根据hash结果将同一个客户端(IP相同)的请求分发给同一台服务器进行处理

例如:不能在每台服务器上都保存客户端的session信息,所以要么就保证同一个客户端的请求每次都发送给特定的服务器,要么就使用redis作为session信息共享
动静分离
关于静态文件
在我们的软件开发中,有些请求是需要后台处理的,有些请求是不需要经过后台处理的(如:css、html、jpg、js等等文件),这些不需要经过后台处理的文件称为静态文件
让动态网站里的动态网页根据一定规则把不变的资源和经常变的资源区分开来,动静资源做好了拆分以后,就可以根据静态资源的特点将其做缓存操作,提高资源响应的速度
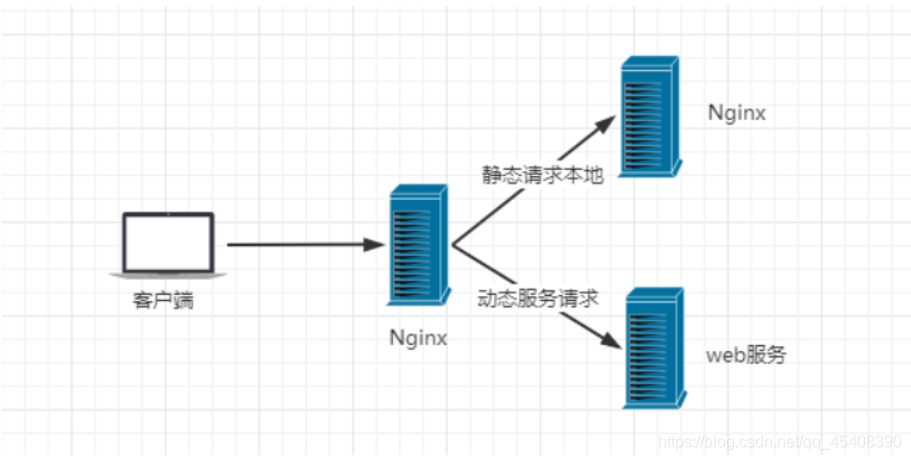
通过使用Nginx进行动静分离,将动态请求转发给后端应用服务器(如PHP-FPM、Node.js等),而将静态请求直接由Nginx服务器处理
Nginx的
Windows下的安装
下载Nginx:http://nginx.org/en/download.html (建议下载稳定版本)
解压完成后:
启动Nginx
方法1:直接双击nginx.exe,双击后一个黑色的弹窗一闪而过
方法2:打开cmd命令窗口,切换到nginx解压目录下,输入命令 nginx.exe ,回车即可

检查Nginx是否启动成功
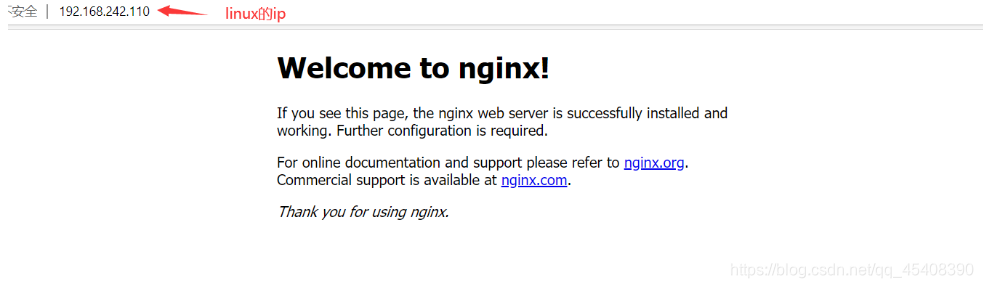
直接在浏览器地址栏输入网址 localhost:80回车,出现以下页面说明启动成功
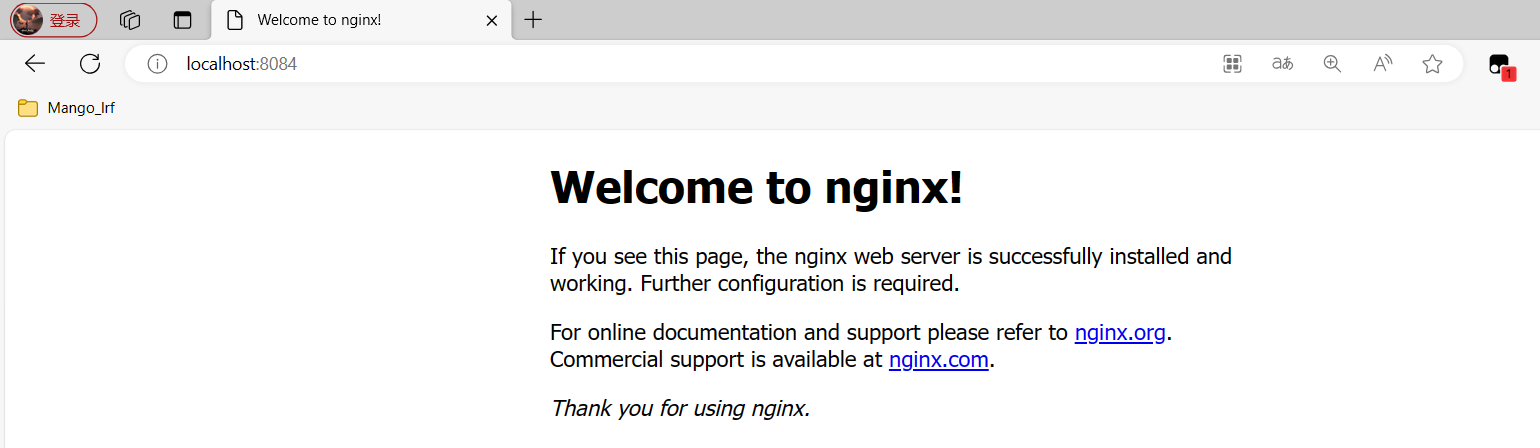
注意:此处我访问的是8084端口,可以在配置文件当中修改nginx监听的端口号
修改配置文件
nginx的配置文件是conf目录下的``nginx.conf`文件,默认配置的nginx监听的端口为80,如果80端口被占用可以修改为未被占用的端口即可
否则启动nginx的时候会出现类似下述的错误,此时只需要修改nginx监听的端口号即可
nginx: [emerg] bind() to 0.0.0.0:80 failed (10013: An attempt was made to access a socket in a way forbidden by its access permissions)
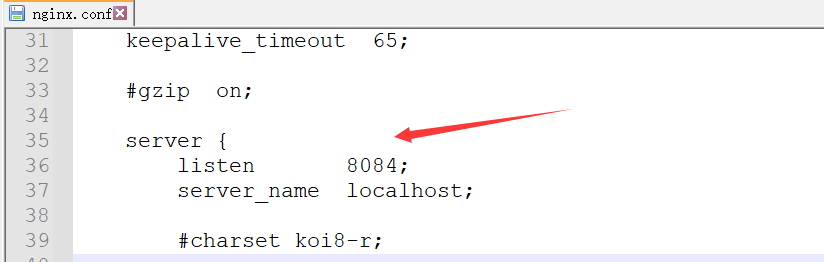
当我们修改了nginx的配置文件nginx.conf 时,不需要关闭nginx后重新启动nginx,只需要在cmd控制台当中执行命令 nginx -s reload 让改动生效
关闭Nginx
如果使用cmd命令窗口启动nginx, 关闭cmd窗口是不能结束nginx进程的,可使用两种方法关闭nginx
方法1:输入nginx命令
nginx -s stop(快速停止nginx)nginx -s quit(完整有序的停止nginx)
方法2:使用taskkill 命令 (taskkill是用来终止进程的)
taskkill /f /t /im nginx.exe- 选项解释:
/f是强制终止/t是终止指定的进程和任何由此启动的子进程,/im是指定进程名称
注意:如果输入命令的时候出现拒绝访问等情况,可以尝试以管理员方式打开cmd控制台
Linux下的安装
前置准备
1)要确定有gcc环境
gcc -v # 查看版本
yum install gcc-c++ # 如果没有的话就安装一下
2)PCRE pcre-devel 安装
PCRE(Perl Compatible Regular Expressions) 是一个Perl库,包括 perl 兼容的正则表达式库
nginx 的 http 模块使用 pcre 来解析正则表达式,所以需要在 linux 上安装 pcre 库,pcre-devel 是使用 pcre 开发的一个二次开发库。nginx也需要此库
yum install -y pcre pcre-devel
3)zlib 安装
zlib 库提供了很多种压缩和解压缩的方式, nginx 使用 zlib 对 http 包的内容进行解压,所以需要在 Centos 上安装 zlib 库。
yum install -y zlib zlib-devel
4)OpenSSL 安装
OpenSSL 是一个强大的安全套接字层密码库,囊括主要的密码算法、常用的密钥和证书封装管理功能及 SSL 协议,并提供丰富的应用程序供测试或其它目的使用。nginx 不仅支持 http 协议,还支持 https,所以需要在 Centos 安装 OpenSSL 库。
yum install -y openssl openssl-devel
5)在官网下载nginx压缩包,并上传到linux上
6)在Linux当中解压安装包
tar -zxvf nginx-1.20.1.tar.gz # 解压安装包
cd nginx-1.20.1/ # 进入目录,发现和windows没什么区别
7)进行自动配置
./configure
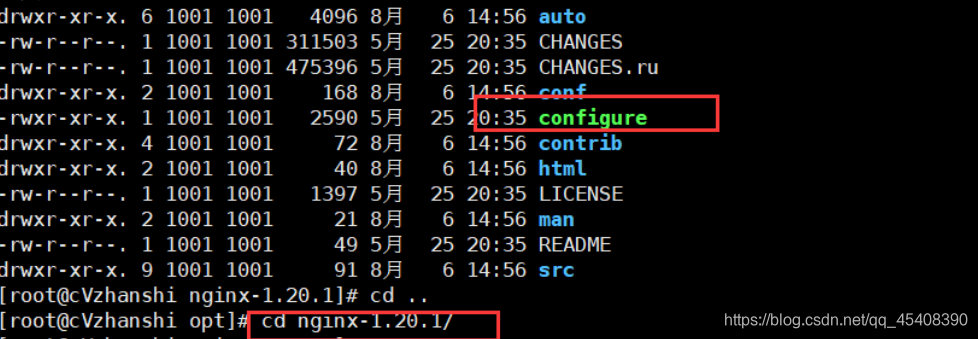
- 执行make命令
- 执行make install命令
- 查看是否安装成功 =>
whereis nginx
补充:whereis 是用于查找可执行文件、源代码文件和帮助文档位置的命令
[root@cVzhanshi nginx-1.20.1]# whereis nginx
nginx: /usr/local/nginx
- 到安装目录去看看
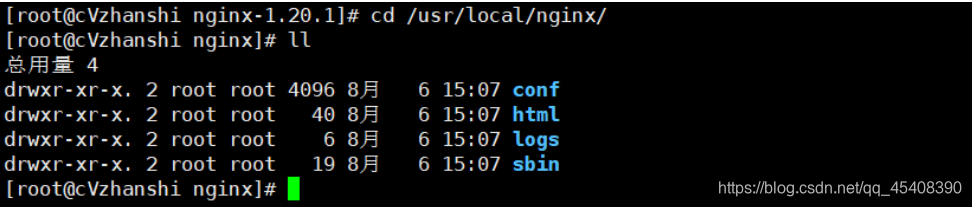
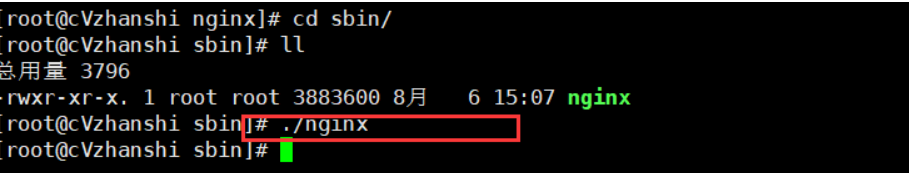
- 进入sbin目录,执行可执行文件
- 在浏览器看是否执行成功
Nginx常用命令
首先需要进入到nginx可执行文件的文件夹当中
cd /usr/local/nginx/sbin/ #进入nginx可执行文件所在的路径
./nginx #启动
./nginx -s stop #强制停止
./nginx -s quit #安全退出
./nginx -s reload #重新加载配置文件 一旦改了配置文件就需要执行该指令才能生效
ps aux|grep nginx #查看nginx进程
注意:如果nginx连接不上,检查阿里云安全组是否开放端口,或者服务器防火墙是否开放端口
防火墙相关命令:
# 开启防火墙
service firewalld start
# 重启
service firewalld restart
# 关闭
service firewalld stop
# 查看防火墙规则
firewall-cmd --list-all
# 查询端口是否开放
firewall-cmd --query-port=8080/tcp
# 开放80端口
firewall-cmd --permanent --add-port=80/tcp
# 移除端口
firewall-cmd --permanent --remove-port=8080/tcp
#重启防火墙(修改配置后要重启防火墙)
firewall-cmd --reload
# 参数解释
1、firwall-cmd:是Linux提供的操作firewall的一个工具;
2、--permanent:表示设置为持久;
3、--add-port:标识添加的端口;
负载均衡功能演示
先进行配置文件的修改
#每一行配置的最后由分号结尾
http {...upstream 任意名字Name{ #负载均衡配置 #服务器资源 (有哪些服务器)#服务器组包含两个服务器,分别是127.0.0.1:8082和127.0.0.1:8081,并且它们的权重都被设置为1server 127.0.0.1:8082/ weight=1; #weight为服务器的权重server 127.0.0.1:8081/ weight=1;}#定义了一个虚拟主机(server),监听默认的80端口,并使用localhost作为服务器名server {#http默认端口号为80 https默认端口号为443listen 80;server_name localhost;#代理转发#只要是80端口 根目录下的请求就代理到http://任意名字Namelocation / {#root指令指定了静态文件的根目录为html,index指令定义了默认的索引文件root html;index index.html index.htm;#proxy_pass指令将请求转发给之前定义的上游服务器组任意名字Name。这意味着当有请求到达根路径时,Nginx会将请求代理转发给上游服务器组中的服务器,实现负载均衡proxy_pass http://任意名字Name;}...
}
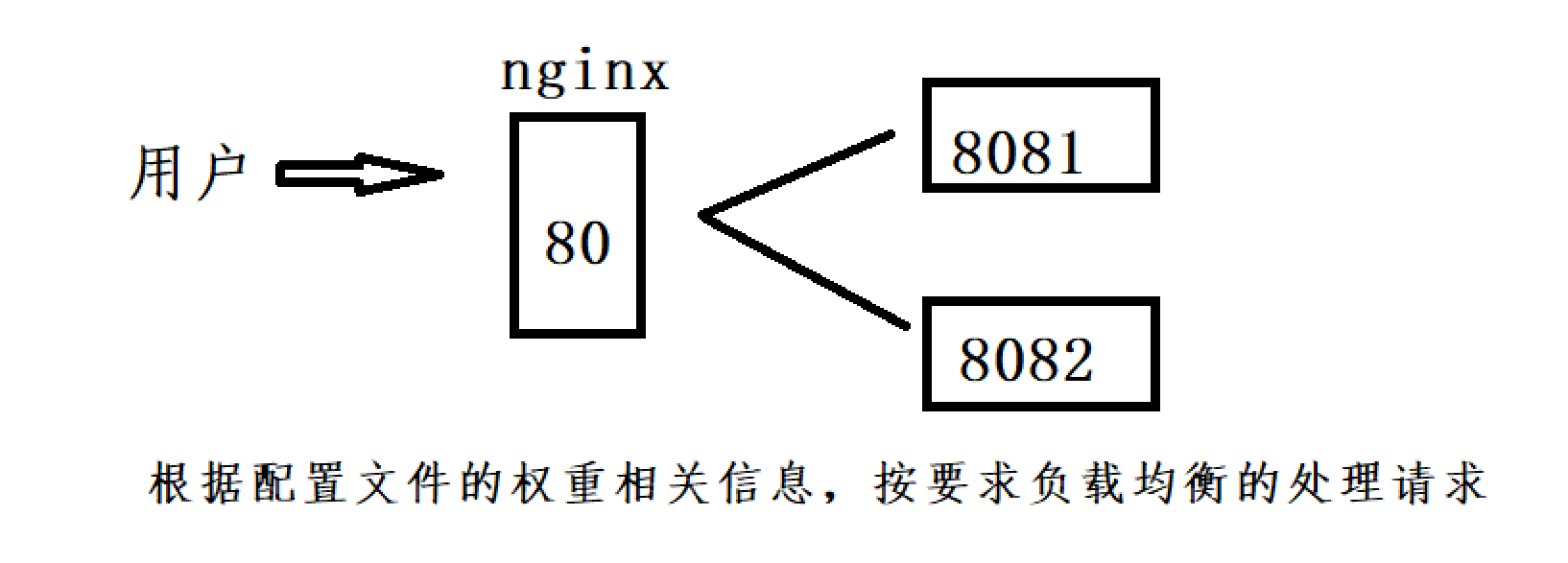
步骤1:启动两个端口的项目
步骤2:启动nginx
步骤3:进行测试